Home
Last updated: 2020-12-28
Checks: 2 0
Knit directory: HowdenWilson2020/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 0a845cc. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/index.Rmd) and HTML (docs/index.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 0a845cc | Sean Wilson | 2020-12-28 | update to flatly theme |
| html | 07adb2a | Sean Wilson | 2020-12-28 | Build site. |
| Rmd | 11477de | Sean Wilson | 2020-12-28 | Updating additional information around analysis |
| html | 1297fee | Sean Wilson | 2020-12-28 | Build site. |
| Rmd | d1d8b3f | Sean Wilson | 2020-12-28 | Initialise website |
| html | 84370d5 | Sean Wilson | 2020-11-13 | Build site. |
| Rmd | 96a2303 | Sean Wilson | 2020-11-09 | Start workflowr project. |
Howden, Wilson et al. 2020
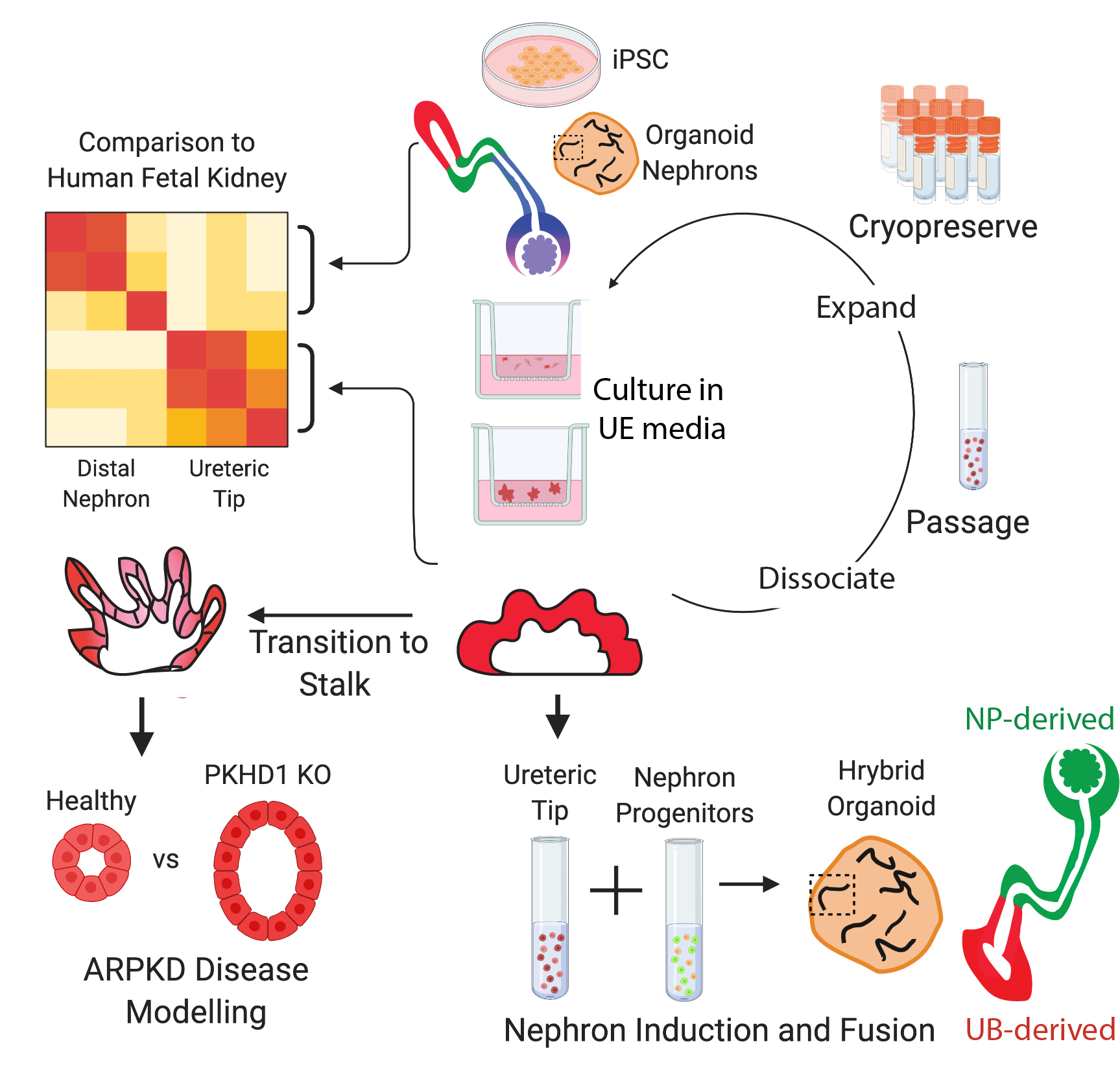
This repository is a workflowr project showing the analysis and output relating to Howden, Wilson et al. 2020: Plasticity of distal nephron epithelia from human kidney organoids enables the induction of ureteric tip and stalk; Cell Stem Cell
Graphical Abstract
Summary
During development, distinct progenitors contribute to the nephrons versus the ureteric epithelium of the kidney. Indeed, previous human pluripotent stem-cell-derived models of kidney tissue either contain nephrons or pattern specifically to the ureteric epithelium. By re-analyzing the transcriptional distinction between distal nephron and ureteric epithelium in human fetal kidney, we show here that, while existing nephron-containing kidney organoids contain distal nephron epithelium and no ureteric epithelium, this distal nephron segment alone displays significant in vitro plasticity and can adopt a ureteric epithelial tip identity when isolated and cultured in defined conditions. “Induced” ureteric epithelium cultures can be cryopreserved, serially passaged without loss of identity, and transitioned toward a collecting duct fate. Cultures harboring loss-of-function mutations in PKHD1 also recapitulate the cystic phenotype associated with autosomal recessive polycystic kidney disease.